Researchers at Gifu University's Institute for Glyco-core Research have developed two synthetic approaches to produce a fragment of a molecule called poly(ADP-ribose). By developing two ways to synthesize the fragment, researchers may be able to better understand how it contributes to vital functions in the cell. Credit: Hiromune Ando and Hide-Nori Tanaka
A sugar-based molecule naturally produced by the body can help cells grow, differentiate into different types, self-destruct if need be and much more. It helps protect the cell's genome, repair DNA, and regulate how genes are passed down. The molecule, called poly(adenosine diphosphate ribose) or poly(ADP-ribose), can potentially inform disease prevention and treatments—if scientists can figure out exactly how it works.
To facilitate such scientific discovery, researchers at Gifu University's Institute for Glyco-core Research (iGCORE) in Japan developed two synthetic versions of an ADP-ribose fragment.
They published their approach in the European Journal of Organic Chemistry.
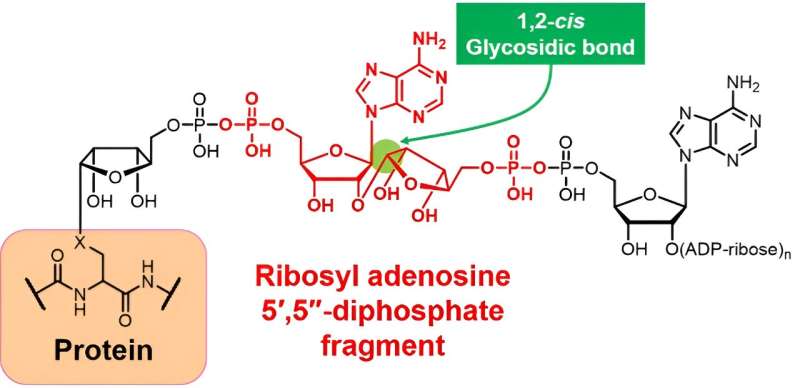
When cells make new proteins, they translate the genetic instructions to machinery that can build the proteins. During that process, some molecules or molecular fragments can bind to the protein as a post-translational modification. The poly(ADP-ribose) fragment, known as ribosyl adenosine 5′,5′′-diphosphate, could help reveal specific cellular functions, but naturally occurring fragments are too varied for scientists to attribute broad functions.
-
Although poly(ADP-ribose) occurs naturally, the molecular fragment—called ribosyl adenosine 5',5"-diphosphate—varies too widely for researchers to infer broad functions. Credit: Hiromune Ando and Hide-Nori Tanaka
-
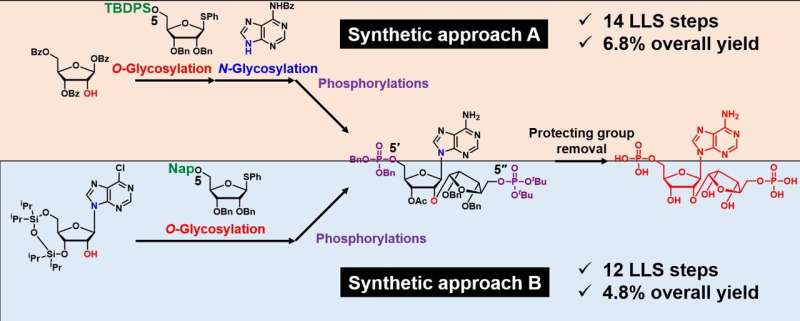
Researchers developed two synthetic approaches to produce the poly(ADP-ribose) fragment. Both approaches may help researchers scale production of more uniform fragments for use in studies aimed at more precisely understanding their role in cellular health. Credit: Hiromune Ando and Hide-Nori Tanaka
"The problem is lack of the availability of homogeneous oligo- and poly(ADP-ribose) samples, which are necessary for molecular-level studies to elucidate their detailed functions," said co-corresponding author Hide-Nori Tanaka, an assistant professor at iGCORE. Oligo- and poly(ADP-ribose) refers to the number of components that bind together to make up the ADP-ribose molecule.
"To address this bottleneck and accelerate ADP-ribose biology, we developed two practical synthetic approaches to ribosyl adenosine 5',5"-diphosphate, a fragment of poly(ADP-ribose), for providing structurally well-defined ADP-ribose oligomer and polymer."
The first method involved a stepwise assembly using a commercially available solution to produce a framework to which the researchers then added carbohydrates. The second approach was streamlined into a single step where researchers processed a known molecule that can bind to other molecules from a commercially available solution. Both methods produced a common precursor that converts into a conjugation-ready building block that is primed for application in ADP-ribose synthesis, according to Tanaka.
"The next step is ADP-ribose oligomer synthesis using the building block we prepared in this paper," Tanaka said. "Our ultimate goal is to elucidate the detailed functions of oligo- and poly(ADP-ribose) by chemical biology approach using synthetic molecules."
More information: Rui Hagino et al, Synthetic Approaches to Ribosyl Adenosine 5′,5′′‐Diphosphate Fragment of Poly(ADP‐ribose), European Journal of Organic Chemistry (2023). DOI: 10.1002/ejoc.202300875
Journal information: European Journal of Organic Chemistry
Provided by Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System